HTML
Short Communication - (2023) Volume 13, Issue 1
Aspects for Evaluating Psychedelic Abuse Risk during Drug Discovery
Jara Natalia**Correspondence: Jara Natalia, Department of Pharmacy, University of Cambridge, Cambridge, United Kingdom, Email:
Received: 02-Jan-2023, Manuscript No. IJP-23-93085; Editor assigned: 05-Jan-2023, Pre QC No. IJP-23-93085 (PQ); Reviewed: 25-Jan-2023, QC No. IJP-23-93085; Revised: 02-Feb-2023, Manuscript No. IJP-23-93085 (R); Published: 10-Feb-2023, DOI: 10.37532/2249-1848.2023.13(1).35
Description
The process of drug design/discovery entails identifying and designing novel molecules with the desired properties and that bind well to a given disease-relevant target. Exploration of the vast drug-like chemical space to find novel chemical structures with desired physiochemical properties and biological characteristics is one of the main challenges in effectively identifying potential drug candidates. Furthermore, the chemical space of currently available molecular libraries represents only a small subset of the total possible drug-like chemical space. Deep molecular generative models have received a lot of attention because they offer an alternative approach to molecule design and discovery. To efficiently explore the drug-like space, we first created a drug-like dataset and then used a Conditional Randomized Transformer (CRT) to generate drug-like molecules. Technique using the Molecular Access System (MACCS) fingerprint as a condition, and compared it to previously reported molecular generative models. The findings reveal that the deep molecular generative model investigates a broader drug-like chemical space.
The drug-like molecules generated share the same chemical space as known drugs, and the drug-like space captured by the combination of Quantitative Estimation of Drug-likeness (QED) and Quantitative Estimation of Protein-Protein Interaction (QEPPI) targeting drug-likeness can cover a larger drug-like space. Medication combination treatment is a potential technique for improving outcomes. The intended therapeutic benefit is achieved while adverse effects are minimized. High-throughput pairwise drug combination screening is a popular approach for uncovering beneficial medication interactions, although it is time-consuming and expensive. We study the application of reaction network topology-guided combination treatment design as a predictive in silico drug-drug interaction screening technique. We concentrated on three-node enzymatic networks with Michaelis-Menten kinetics. The findings demonstrated that drug-drug interactions are heavily influenced by the target arrangement chosen in a specific topology, the type of the drug, and the intended amount of change in network output. The findings revealed a negative relationship between antagonistic interactions and medication dose. Generally, negative feedback loops had the most synergistic. It, somewhat surprisingly, needed the largest medication dosages when compared to other topologies under the same conditions. The medication delivery system distributes pharmaceuticals to the target spot for effectiveness while avoiding damaging side effects on normal cells. The goal of a drug delivery system is to prevent harmful side effects, improve bioavailability, limit degradation, maintain steady and effective blood concentration, avoid blood concentration variations, and raise drug concentration in target tissues. To build drug delivery systems, two strategies are used: one is to use delivery carriers to improve drug stability, and the other is a prodrug strategy in which the drug is covalently modified to temporarily limit its activity. In comparison to the former, the prodrug method eliminates the safety difficulties of immunogenicity and toxicity induced by delivery carriers and minimizes patients' metabolic load on delivery carriers, and hence offers a wide variety of applications.
Peptide Drug Conjugates (PDCs) are pharmaceuticals chemically bonded to peptide sequences with specific functionalities via specialized linkers and are considered a new prodrug approach in targeted drug delivery systems. PDCs have an intrinsic chemical structure constituted of three components: peptide, linker, and drug, the mechanism of action of which varies depending on the type of peptides and linkers used. To begin, peptides precisely target cells by binding to receptors on their surfaces, and PDCs enter the cell via receptor-mediated internalization. The linker then disintegrates in the cell in response to stimulation, allowing the protein to be released. A prodrug method like this has the potential to enhance drug targeting, decrease harmful side effects on other cells, boost medication stability in blood circulation, regulate drug release, and improve drug bioavailability.
Conclusion
As compared to Antibody–Drug Conjugates (ADCs) with comparable building processes, the peptides in PDC offer distinct benefits. Certain targeting peptides can reduce drug resistance in tumor cells by modifying the cell entrance mechanism, resulting in efficient killing of drug-resistant tumors and breaking the cycle of unsuccessful therapy owing to drug resistance, which is widespread in conventional chemotherapy. PDCs' short peptide properties make their structures more flexible and easier to modify and conjugate, allowing them to be combined with a variety of drugs, including chemical, protein, and peptide drugs, to prepare targeted agents, significantly reducing off-target toxicity and greatly improving the feasibility of PDC formulation platform technology. Peptide fragment synthesis is straightforward and easy to scale up. As a result, PDC offers enormous promise for designing tailored medication delivery systems.
References
- Babaei M, Evers TM, Shokri F, et al. Comput Biol Med. 2023;155:106584.
- Gong L, Zhao H, Liu Y, et al. Acta Pharm Sin B. 2023.
- Wang J, Mao J, Wang M, et al. Methods. 2023.
- Calderon SN, Bonson KR, Reissig CJ, et al. Neuropharmacology. 2022:109352.
- Giombi K, Thompson J, Wines C, et al. Res Social Adm Pharm. 2023.
- Harris MS, Howden CW, Tomaino J, et al. Gastroenterology. 2020;158(1):18-21.
- Paras ML, Wolfe SB, Bearnot B, et al. J Thorac Cardiovasc Surg. 2021.
- Ahmad J, Odin JA, Hayashi PH, et al. Drug Alcohol Depend. 2021;218:108426.
- Black TA, Buchwald UK. J Clin Tuberc Other Mycobact Dis. 2021;25:100285.
- Patel D, Zhang Y, Dong Y, et al. J Control Release. 2021;333:65-75.
- Dedroog S, Boel E, Kindts C, et al. Int J Pharm. 2021;609:121201.
Manuscript Submission
Submit your manuscript at Online Submission System
Google scholar citation report
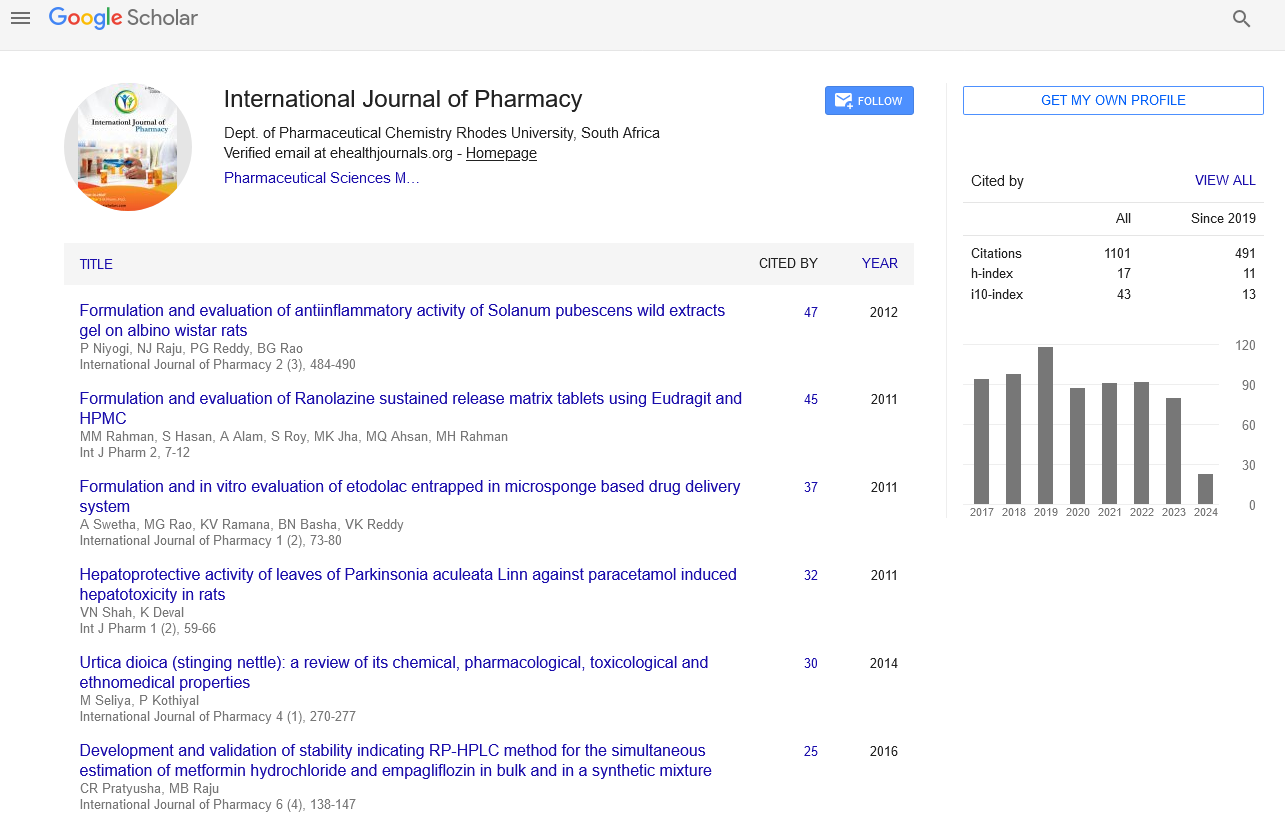
Citations : 1101
International Journal of Pharmacy received 1101 citations as per google scholar report
International Journal of Pharmacy peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- HINARI
- Index Copernicus
- Google Scholar
- The Global Impact Factor (GIF)
- Polish Scholarly Bibliography (PBN)
- Cosmos IF
- Open Academic Journals Index (OAJI)
- Directory of Research Journal Indexing (DRJI)
- EBSCO A-Z
- OCLC- WorldCat
- MIAR
- International committee of medical journals editors (ICMJE)
- Scientific Indexing Services (SIS)
- Scientific Journal Impact Factor (SJIF)
- Euro Pub
- Eurasian Scientific Journal Index
- Root indexing
- International Institute of Organized Research
- InfoBase Index
- International Innovative Journal Impact Factor
- J-Gate